Growth Models¶
The models module contains functions for fitting and selecting growth models to growth curve data.
This module contains:
fit_model()is the main function of the model; it fits growth models to growth curve data.Growth functions (
*_function) are Python function in which the first argumet is time and the rest of the argument are model parameters. Time can be given an anumpy.ndarray.Growth models (
*_model) arelmfit.model.Modelobjects from the lmfit package. These objects wrap the growth functions and are used to fit models to data with parameter constraints and some useful statistics and models selection methods.Parameter guessing functions (
guess_*) are function that use the shape of a growth curve to guess growth parameters without fitting a model to the data, but rather through analytic properties of the growth functions.Hypothesis testing (
has_*) are functions that perform a statistical test to decide if a property of one model can be significantly observed in the data.Outlier functions (
find_outliers*) are functions for identification of outlier growth curve in high-throughput data sets.Indirect trait calculation (
find_*) are functions for calculating growth parameters that are indirectly represented by the models.and some other utilities.
Models¶
Curveball uses the logistic model and its derivaties, the Richards model and the Baranyi-Roberts model:
Logistic model¶
Also known as the Valhulst model, the logistic model includes three parameters and is the simplest and most commonly used population growth model.
The logistic model is defined by an ordinary differential equation (ODE) which also has a closed form solution:
N: population size
r: initial per capita growth rate
K: maximum population size
Richards model¶
Also known as the Generalised logistic model,
Richards model [Richards1959] (or in its discrete time version, the
r: initial per capita growth rate
K: maximum population size
The logistic model is then a special case of the Richards model for
When
When
Baranyi-Roberts model¶
The Baranyi-Roberts model [Baranyi1994] extends Richards model by introducing a lag phase
in which the population growth is slower than expected while it is adjusting to a new environment.
This extension is done by introducing and adjustment function,
where
The model is then described by the following ODE and exact solution:
r: initial per capita growth rate
K: maximum population size
v: adjustment rate
Note that
Richards model is then a special case of the Baranyi-Roberts model for
Logistic models with lag phase¶
We define tow additional models: Baranyi-Roberts with
Model hierarchy and the likelihood ratio test¶
The nesting hierarchy between the models is given in Fig. 1.
This nesting is used for the Likelihood ratio test using the function curveball.models.lrtest()
in which the nested model (to which the arrows point) is the first argument m0,
and the nesting model (from which the arrow points) is the second argument m1.
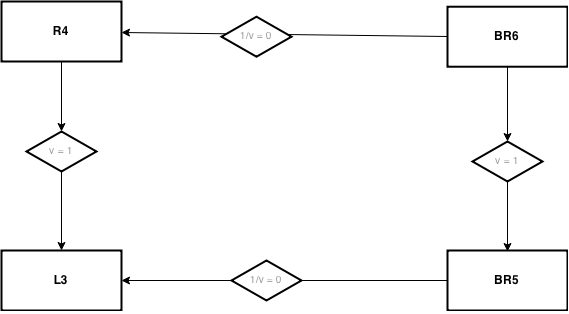
Fig. 6 Fig. 1 - Map of model nesting for the likelihood ratio test.¶
Parameter guessing¶
Before fitting models to the data, Curveball attempts to guess the growth parameters from the shape of the curve. These guesses are used as initial parameters in the model fitting procedure.
y0 and K are guessed from the minimum and maximum of the growth curve, respectively.
r and nu are guessed in guess_r() and guess_nu(), respectively, using formulas from [Richards1959]:
r: initial per capita growth rate
K: maximum population size
q0 and v, the lag phase parameters, are guessed by fitting a Baranyi-Roberts model with fixed
y0, r, K, and nu (based on guesses) to the data.
Model selection¶
Model selection is done by comparing the Bayesian Information Criteria (BIC)
of all model fits and choosing the model fit with the lowest BIC value.
BIC is a common method to measure the quality of a model fit [Kaas1995],
balancing between model fit (distance from data) and complexity (number of parameters).
See lmfit.model.ModelFit.bic() for more information.
Curveball also calculates the weighted BIC each model fitted to the same data. This can be interpreted as the weight of evidence for each model.
Example¶
>>> import pandas as pd
>>> import curveball
>>> plate = pd.read_csv('plate_templates/G-RG-R.csv')
>>> df = curveball.ioutils.read_tecan_xlsx('data/Tecan_280715.xlsx', label='OD', plate=plate)
>>> models, fig, ax = curveball.models.fit_model(df[df.Strain == 'G'], PLOT=True, PRINT=False)
Fig. 7 Fig. 2 - Model fitting and selection. The figure is the result of calling fig.savefig() on the result of the above example code.
Red error bars: mean and standard deviation of data; Green solid line: model fit; Blue dashed line: initial guess.¶
References¶
Richards, F. J., 1959. A Flexible Growth Function for Empirical Use. Journal of Experimental Botany
Baranyi, J., Roberts, T. A., 1994. A dynamic approach to predicting bacterial growth in food. Int. J. Food Microbiol.
Kass, R., Raftery, A., 1995. Bayes Factors. J. Am. Stat. Assoc.
Gilpin, M. E., Ayala, F. J., 1973. Global Models of Growth and Competition. Proc. Nat. Acad. Sci. U S A.
Baty, Florent, and Marie-Laure Delignette-Muller. 2004. Estimating the Bacterial Lag Time: Which Model, Which Precision?. Intl. J. Food Microbiol.
Members¶
- curveball.models.bootstrap_params(df, model_result, nsamples, unit='Well', fit_kws=None)[source]¶
Sample model parameters by fitting the model to resampled data.
The data is bootstraped by drawing growth curves (sampling from the unit column in df) with replacement.
Parameters¶
- dfpandas.DataFrame
the data to fit
- model_resultlmfit.model.ModelResult
result of fitting a
lmfit.model.Modelto data indf- nsamplesint
number of samples to draw
- unitstr, optional
the name of the column in df that identifies a resampling unit, defaults to
Well- fit_kwsdict, optional
dict of kwargs for fit_model
Returns¶
- pandas.DataFrame
data frame of samples; each row is a sample, each column is a parameter.
Raises¶
ValueError : if model_result isn’t an instance of
lmfit.model.ModelResultValueError : if df is emptySee also¶
sample_params
- curveball.models.calc_weights(df, PLOT=False)[source]¶
Calculate weights for the fittiing procedure based on the standard deviations at each time point.
If there is more than one replicate, use the standard deviations as weight. Warn about NaN and infinite values.
Parameters¶
- dfpandas.DataFrame
data frame with Time and OD columns
- PLOTbool, optional
if
True, plot the weights by time
Returns¶
- weightsnp.ndarray
array of weights, calculated as the inverse of the standard deviations at each time point
- figmatplotlib.figure.Figure
if the argument PLOT was
True, the generated figure.- axmatplotlib.axes.Axes
if the argument PLOT was
True, the generated axis.
- curveball.models.cooks_distance(df, model_fit, use_weights=True)[source]¶
Calculates Cook’s distance of each well given a specific model fit.
Cook’s distance is an estimate of the influence of a data curve when performing model fitting; it is used to find wells (growth curve replicates) that are suspicious as outliers. The higher the distance, the more suspicious the curve.
Parameters¶
- dfpandas.DataFrame
growth curve data, see
curveball.ioutilsfor a detailed definition.- model_fitlmfit.model.ModelResult
result of model fitting procedure
- use_weightsbool, optional
should the function use standard deviation across replicates as weights for the fitting procedure, defaults to
True.
Returns¶
- dict
a dictionary of Cook’s distances: keys are wells (from the Well column in df), values are Cook’s distances.
Notes¶
- curveball.models.derivative(func, x0, dx=1.0, n=1, order=3)[source]¶
Finite-difference derivative fallback for SciPy>=1.10 compatibility.
- curveball.models.find_K_ci(param_samples, ci=0.95)[source]¶
Estimates a confidence interval for
K, the maximum population density from the model fit.The confidence interval of the doubling time is the lower and higher percentiles such that ci percent of the max densities are within the confidence interval.
Parameters¶
- param_samplespandas.DataFrame
parameter samples, generated using
sample_params()orbootstrap_params()- cifloat, optional
the fraction of doubling times that should be within the calculated limits. 0 < ci <, defaults to 0.95.
Returns¶
- low, highfloat
the lower and the higher boundaries of the confidence interval of the maximum population density.
- curveball.models.find_all_outliers(df, model_fit, deviations=2, max_outlier_fraction=0.1, use_weights=True, PLOT=False)[source]¶
Iteratively find outlier wells in growth curve data.
At each iteration, calls
find_outliers(). Iterations stop when no more outliers are found or when the fraction of wells defined as outliers is above max_outlier_fraction.Parameters¶
- dfpandas.DataFrame
growth curve data, see
curveball.ioutilsfor a detailed definition.- model_fitlmfit.model.ModelResult
result of model fitting procedure
- deviationsfloat, optional
the number of standard deviations that defines an outlier, defaults to 2.
- max_outlier_fractionfloat, optional
maximum fraction of wells to define as outliers, defaults to 0.1 = 10%.
- use_weightsbool, optional
should the function use standard deviation across replicates as weights for the fitting procedure, defaults to
True.- axmatplotlib.axes.Axes
an axes to plot into; if not provided, a new one is created.
- PLOTbool, optional
if
True, the function will plot the Cook’s distances of the wells and the threshold.
Returns¶
- outlierslist
a list of lists: the list nested at index i contains the labels of the outlier wells found at iteration i.
- figmatplotlib.figure.Figure
if the argument PLOT was
True, the generated figure.- axmatplotlib.axes.Axes
if the argument PLOT was
True, the generated axis.
- curveball.models.find_lag(model_fit, params=None)[source]¶
Estimates the lag duration from the model fit.
The function calculates the tangent line to the model curve at the point of maximum derivative (the inflection point). The time when this line intersects with
Parameters¶
- model_fitlmfit.model.ModelResult
the result of a model fitting procedure
- paramslmfit.parameter.Parameters, optional
if provided, these parameters will override model_fit’s parameters
Returns¶
- lamfloat
the lag phase duration in the units of the model_fit
Timevariable (usually hours).
References¶
[fig2.2]Fig. 2.2 pg. 19 in Baranyi, J., 2010. Modelling and parameter estimation of bacterial growth with distributed lag time..
See also¶
find_lag_ci has_lag
- curveball.models.find_lag_ci(model_fit, param_samples, ci=0.95)[source]¶
Estimates a confidence interval for the lag duration from the model fit.
The lag duration for each parameter sample is calculated. The confidence interval of the lag is the lower and higher percentiles such that ci percent of the random lag durations are within the confidence interval.
Parameters¶
- model_fitlmfit.model.ModelResult
the result of a model fitting procedure
- param_samplespandas.DataFrame
parameter samples, generated using
sample_params()orbootstrap_params()- cifloat, optional
the fraction of lag durations that should be within the calculated limits. 0 < ci <, defaults to 0.95.
Returns¶
- low, highfloat
the lower and the higher boundaries of the confidence interval of the lag phase duration in the units of the model_fit
Timevariable (usually hours).
See also¶
find_lag has_lag
- curveball.models.find_max_growth(model_fit, params=None, after_lag=True)[source]¶
Estimates the maximum population and specific growth rates from the model fit.
The function calculates the maximum population growth rate
For example, in the logistic model the population growth rate is a quadratic function of
In contrast, the per capita growth rate a linear function of
Parameters¶
- model_fitlmfit.model.ModelResult
the result of a model fitting procedure
- paramslmfit.parameter.Parameters, optional
if provided, these parameters will override model_fit’s parameters
- after_lagbool
if true, only explore the time after the lag phase. Otherwise start at time zero. Defaults to
True.
Returns¶
- t1float
the time when the maximum population growth rate is achieved in the units of the model_fit
Timevariable.- y1float
the population size or density (OD) for which the maximum population growth rate is achieved.
- afloat
the maximum population growth rate.
- t2float
the time when the maximum per capita growth rate is achieved in the units of the model_fit Time variable.
- y2float
the population size or density (OD) for which the maximum per capita growth rate is achieved.
- mufloat
the the maximum specific (per capita) growth rate.
See also¶
find_max_growth_ci
- curveball.models.find_max_growth_ci(model_fit, param_samples, after_lag=True, ci=0.95)[source]¶
Estimates a confidence interval for the maximum population/specific growth rates from the model fit.
The maximum population/specific growth rate for each parameter sample is calculated. The confidence interval of the rate is the lower and higher percentiles such that ci percent of the random rates are within the confidence interval.
Parameters¶
- model_fitlmfit.model.ModelResult
the result of a model fitting procedure
- param_samplespandas.DataFrame
parameter samples, generated using
sample_params()orbootstrap_params()- after_lagbool
if true, only explore the time after the lag phase. Otherwise start at time zero. Defaults to
True- cifloat, optional
the fraction of lag durations that should be within the calculated limits. 0 < ci <, defaults to 0.95
Returns¶
- low_a, high_afloat
the lower and the higher boundaries of the confidence interval of the the maximum population growth rate in the units of the model_fit
OD/Time(usually OD/hours).- low_mu, high_mufloat
the lower and the higher boundaries of the confidence interval of the the maximum specific growth rate in the units of the model_fit 1/
Timevariable (usually 1/hours).
See also¶
find_max_growth
- curveball.models.find_min_doubling_time(model_fit, params=None, PLOT=False)[source]¶
Estimates the minimal doubling time from the model fit.
The function evaluates a growth curves based on the model fit and supplied parameters (if any) and calculates that time required to double the population density at each time point. It then returns the minimal doubling time.
Parameters¶
- model_fitlmfit.model.ModelResult
the result of a model fitting procedure
- paramslmfit.parameter.Parameters, optional
if provided, these parameters will override model_fit’s parameters
- PLOTbool, optional
if
True, the function will plot the Cook’s distances of the wells and the threshold. # FIXME
Returns¶
- min_dblfloat
the minimal time it takes the population density to double.
- figmatplotlib.figure.Figure
if the argument PLOT was
True, the generated figure.- axmatplotlib.axes.Axes
if the argument PLOT was
True, the generated axis.
- curveball.models.find_min_doubling_time_ci(model_fit, param_samples, ci=0.95)[source]¶
Estimates a confidence interval for the minimal doubling time from the model fit.
The minimal doubling time for each parameter sample is calculated. The confidence interval of the doubling time is the lower and higher percentiles such that ci percent of the random lag durations are within the confidence interval.
Parameters¶
- model_fitlmfit.model.ModelResult
the result of a model fitting procedure
- param_samplespandas.DataFrame
parameter samples, generated using
sample_params()orbootstrap_params()- cifloat, optional
the fraction of doubling times that should be within the calculated limits. 0 < ci <, defaults to 0.95.
Returns¶
- low, highfloat
the lower and the higher boundaries of the confidence interval of the minimal doubling times in the units of the model_fit
Timevariable (usually hours).
See also¶
find_min_doubling_time
- curveball.models.find_outliers(df, model_fit, deviations=2, use_weights=True, ax=None, PLOT=False)[source]¶
Find outlier wells in growth curve data.
Uses the Cook’s distance approach (cooks_distance); values of Cook’s distance that are deviations standard deviations above the mean are defined as outliers.
Parameters¶
- dfpandas.DataFrame
growth curve data, see
curveball.ioutilsfor a detailed definition.- model_fitlmfit.model.ModelResult
result of model fitting procedure
- deviationsfloat, optional
the number of standard deviations that defines an outlier, defaults to 2.
- use_weightsbool, optional
should the function use standard deviation across replicates as weights for the fitting procedure, defaults to
True.- axmatplotlib.axes.Axes, optional
an axes to plot into; if not provided, a new one is created.
- PLOTbool, optional
if
True, the function will plot the Cook’s distances of the wells and the threshold.
Returns¶
- outlierslist
the labels of the outlier wells.
- figmatplotlib.figure.Figure
if the argument PLOT was
True, the generated figure.- axmatplotlib.axes.Axes
if the argument PLOT was
True, the generated axis.
- curveball.models.fit_exponential_growth_phase(t, N, k=2)[source]¶
Fits an exponential model to the exponential growth phase.
Fits a polynomial p(t)~N, finds tmax the time of the maximum of the derivative dp/dt, and fits a linear function to log(N) around tmax. The resulting slope (a) and intercept (b) are the parameters of the exponential model:
Arguments¶
- tnp.ndarray
time
- Nnp.ndarray
N[i] is the population size at time t[i]
- kint
number of points to take around tmax, defaults to 2 for a total of 5 points
Returns¶
- slopefloat
slope of the linear regression, a
- interceptfloat
intercept of the linear regression, b
- curveball.models.fit_model(df, param_guess=None, param_min=None, param_max=None, param_fix=None, models=None, use_weights=False, use_Dfun=False, method='leastsq', ax=None, PLOT=True, PRINT=True)[source]¶
Fit and select a growth model to growth curve data.
This function fits several growth models to growth curve data (
ODas a function ofTime).Parameters¶
- dfpandas.DataFrame
growth curve data, see
curveball.ioutilsfor a detailed definition.- param_guessdict, optional
a dictionary of parameter guesses to use (key:
strof param name; value:floatof param guess).- param_mindict, optional
a dictionary of parameter minimum bounds to use (key:
strof param name; value:floatof param min bound).- param_maxdict, optional
a dictionary of parameter maximum bounds to use (key:
strof param name; value:floatof param max bound).- param_fixset, optional
a set of names (
str) of parameters to fix rather then vary, while fitting the models.- use_weightsbool, optional
should the function use the deviation across replicates as weights for the fitting procedure, defaults to
False.- use_Dfunbool, optional
should the function calculate the partial derivatives of the model functions to be used in the fitting procedure, defaults to
False.- modelsone or more model classes, optional
model classes (not instances) to use for fitting; defaults to all model classes in curveball.baranyi_roberts_model.
- methodstr, optional
the minimization method to use, defaults to leastsq, can be anything accepted by
lmfit.minimizer.Minimizer.minimize()orlmfit.minimizer.Minimizer.scalar_minimize().- axmatplotlib.axes.Axes, optional
an axes to plot into; if not provided, a new one is created.
- PLOTbool, optional
if
True, the function will plot the all model fitting results.- PRINTbool, optional
if
True, the function will print the all model fitting results.
Returns¶
- modelslist of lmfit.model.ModelResult
all model fitting results, sorted by increasing BIC (a measure of fit quality).
- figmatplotlib.figure.Figure
figure object.
- axnumpy.ndarray
array of
matplotlib.axes.Axesobjects, one for each model result, with the same order as models.
Raises¶
- TypeError
if one of the input parameters is of the wrong type (not guaranteed).
- ValueError
if the input is bad, for example, df is empty (not guaranteed).
- AssertionError
if any of the intermediate calculated values are inconsistent (for example,
y0<0).
Examples¶
>>> import curveball >>> import pandas as pd >>> import matplotlib.pyplot as plt >>> plate = pd.read_csv('plate_templates/G-RG-R.csv') >>> df = curveball.ioutils.read_tecan_xlsx('data/Tecan_280715.xlsx', label='OD', plate=plate) >>> green_models = curveball.models.fit_model(df[df.Strain == 'G'], PLOT=True)
- curveball.models.get_models(module)[source]¶
Finds and returns all models in module.
Parameters¶
- modulemodule
the module in which to look for models
Returns¶
- list
list of subclasses of
lmfit.model.Modelthat can be used withcurveball.models.fit_model()
See also¶
fit_model is_model curveball.baranyi_roberts_model
- curveball.models.has_lag(model_fits, alfa=0.05, PRINT=False)[source]¶
Checks if if the best fit has statisticaly significant lag phase
If the best fitted model doesn’t has a lag phase to begin with, return
False. This includes the logistic model and Richards model.Otherwise, a likelihood ratio test will be perfomed with nesting determined according to Figure 1. The null hypothesis of the test is that
The function will return
Trueif the null hypothesis is rejected, otherwise it will returnFalse.Parameters¶
- model_fitssequence of lmfit.model.ModelResult
the results of several model fitting procedures, ordered by their statistical preference. Generated by
fit_model().- alfafloat, optional
test significance level, defaults to 0.05 = 5%.
- PRINTbool, optional
if
True, the function will print the result of the underlying statistical test; defaults toFalse.
Returns¶
- bool
the result of the hypothesis test.
Trueif the null hypothesis was rejected and the data suggest that there is a significant lag phase.
Raises¶
- ValueError
raised if the fittest of the
lmfit.model.ModelResultobjects in model_fits is of an unknown model.
- curveball.models.has_nu(model_fits, alfa=0.05, PRINT=False)[source]¶
Checks if if the best fit has
If the best fitted model has
False. This includes the logistic model. Otherwise, a likelihood ratio test will be perfomed with nesting determined according to Figure 1. The null hypothesis of the test is thatTrue. Otherwise it will returnFalse.Parameters¶
- model_fitslist lmfit.model.ModelResult
the results of several model fitting procedures, ordered by their statistical preference. Generated by
fit_model().- alfafloat, optional
test significance level, defaults to 0.05 = 5%.
- PRINTbool, optional
if
True, the function will print the result of the underlying statistical test; defaults toFalse.
Returns¶
- bool
the result of the hypothesis test.
Trueif the null hypothesis was rejected and the data suggest that
Raises¶
- ValueError
raised if the fittest of the
lmfit.model.ModelResultobjects in model_fits is of an unknown model.
- curveball.models.information_criteria_weights(results)[source]¶
Calculate weighted AIC and BIC for model results.
where
Parameters¶
- resultssequence of lmfit.model.ModelResult
use the
lmfit.attr.ModelResult.aicandlmfit.model.ModelResult.bicattributes to add a weighted_aic and weighted_bic attribute.
Notes¶
- curveball.models.is_model(cls)[source]¶
Returns
Trueif the input is a subclass oflmfit.model.Model.Parameters¶
cls : class
Returns¶
bool
- curveball.models.lrtest(m0, m1, alfa=0.05)[source]¶
Performs a likelihood ratio test on two nested models.
For two models, one nested in the other (meaning that the nested model estimated parameters are a subset of the nesting model), the test statistic
where
The function compares between two
lmfit.model.ModelResultobjects. These are the results of fitting models to the same dataset using the lmfit package.The function compares between model fit m0 and m1 and assumes that m0 is nested in m1, meaning that the set of varying parameters of m0 is a subset of the varying parameters of m1.
lmfit.model.ModelResult.chisqris the sum of the square of the residuals of the fit.lmfit.model.ModelResult.ndatais the number of data points.lmfit.model.ModelResult.nvarysis the number of varying parameters.Parameters¶
- m0, m1lmfit.model.ModelResult
objects representing two model fitting results. m0 is assumed to be nested in m1.
- alfafloat, optional
test significance level, defaults to 0.05 = 5%.
Returns¶
- prefer_m1bool
should we prefer m1 over m0
- pvalfloat
the test p-value
- Dfloat
the test statistic
- ddfint
the number of degrees of freedom
Notes¶
- curveball.models.sample_params(model_fit, nsamples, params=None, covar=None)[source]¶
Random sample of parameter values from a truncated multivariate normal distribution defined by the covariance matrix of the a model fitting result.
Parameters¶
- model_fitlmfit.model.ModelResult
the model fit that defines the sampled distribution
- nsamplesint
number of samples to make
- paramsdict, optional
a dictionary of model parameter values; if given, overrides values from model_fit
- covarnumpy.ndarray, optional
an array containing the parameters covariance matrix; if given, overrides values from model_fit
Returns¶
- pandas.DataFrame
data frame of samples; each row is a sample, each column is a parameter.
See also¶
bootstrap_params